The obvious discrepancy between the relative genome-wide mutation rates and relative synonymous site divergences may be at least partly explained by the difference in base composition between the mitochondrial genome as a complete and its synonymous websites. Mitochondrial synonymous websites are extremely A+T-rich and so are expected to mutate at a decrease frequency than the mitochondrial genome as a complete, which is according to the low frequency of synonymous mutations that we observed (Table 3). Our excessive mitochondrial mutation rate estimate largely comes from mutations at nonsynonymous major-strand G websites; these are topic to robust purifying selection in nature, and this contribute little to between-species divergence. Molecular clock users have developed workaround solutions utilizing numerous statistical approaches including maximum likelihood strategies and later Bayesian modeling. In explicit, models that bear in mind price variation throughout lineages have been proposed so as to acquire better estimates of divergence times.
Save citation to file
the culturing of the cells, the researchers subsequent isolated DNA from the original
For most operators (like random walk and subtree slide operators) a larger tuning parameter means larger strikes. However for the dimensions operator a tuning parameter value closer to 0.0 means greater moves. At the highest of the window is an option called Auto Optimize which, when chosen, will automatically regulate the tuning setting as the MCMC runs to try to achieve most effectivity.
Create a file for exterior citation administration software
In each knowledge sets, the mutation rate was considerably variable throughout haplogroups (see additionally, supplementary fig. S10, Supplementary Material online). (B and D) Variation in somatic mutation rate is correlated with department length heterogeneity in the 1KG (B) and HGDP (D) knowledge units, suggesting that interhaplogroup mutation rate variation is a parsimonious explanation for branch length heterogeneity. In people and different species, pedigree evaluation has suggested a considerably higher mitochondrial mutation fee than the speed indirectly inferred from between-species phylogenetic comparisons [4,27]. The human mitochondrial genome as a whole and the control region are much less biased in their composition than D.
Molecular-clock strategies for estimating evolutionary charges and timescales
For example, assuming that greater mutation rate is ancestral, there have been probably multiple slowdown events which occurred independently within the ancestors of haplogroups E and R. Our conclusions were unlikely driven by batch results (supplementary observe four, Supplementary Material online). In summary, our findings indicate that there is substantial interhaplogroup variation in Y-chromosome mutation fee, and that such variation is a parsimonious explanation for phylogenetic department size heterogeneity. We assumed that mutations seem within the mitochondrial genome at a price μ per site per era, that μ is sufficiently low that a number of mutation occasions on the same site can be ignored, and that the fates of recent mutations are decided solely by genetic drift. Under a neutral model, the fixation rate at equilibrium between drift and mutation is 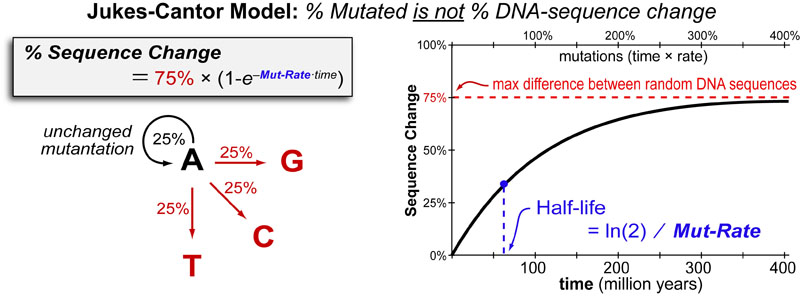 proportional to the mutation rate [13].
proportional to the mutation rate [13].
Even with an correct topology, rate variation can bias the estimate of divergence occasions with molecular clock based strategies. For this cause, previous research of substitution rate variation in plant mitochondrial genomes have constrained their analyses primarily based on phylogenies and divergence instances inferred from nuclear and chloroplasts sequences. Evolutionary genetics research human historical past within a chronological molecular context.